[(Cp-R)M(CO)(3) ] (M=Re or (99m) Tc) Arylsulfonamide, Arylsulfamide, and Arylsulfamate Conjugates for Selective Targeting of Human Carbonic Anhydrase IX.
Can, D., Spingler, B., Schmutz, P., Mendes, F., Raposinho, P., Fernandes, C., Carta, F., Innocenti, A., Santos, I., Supuran, C.T., Alberto, R.(2012) Angew Chem Int Ed Engl 51: 3354-3357
- PubMed: 22344779
- DOI: https://doi.org/10.1002/anie.201107333
- Primary Citation of Related Structures:
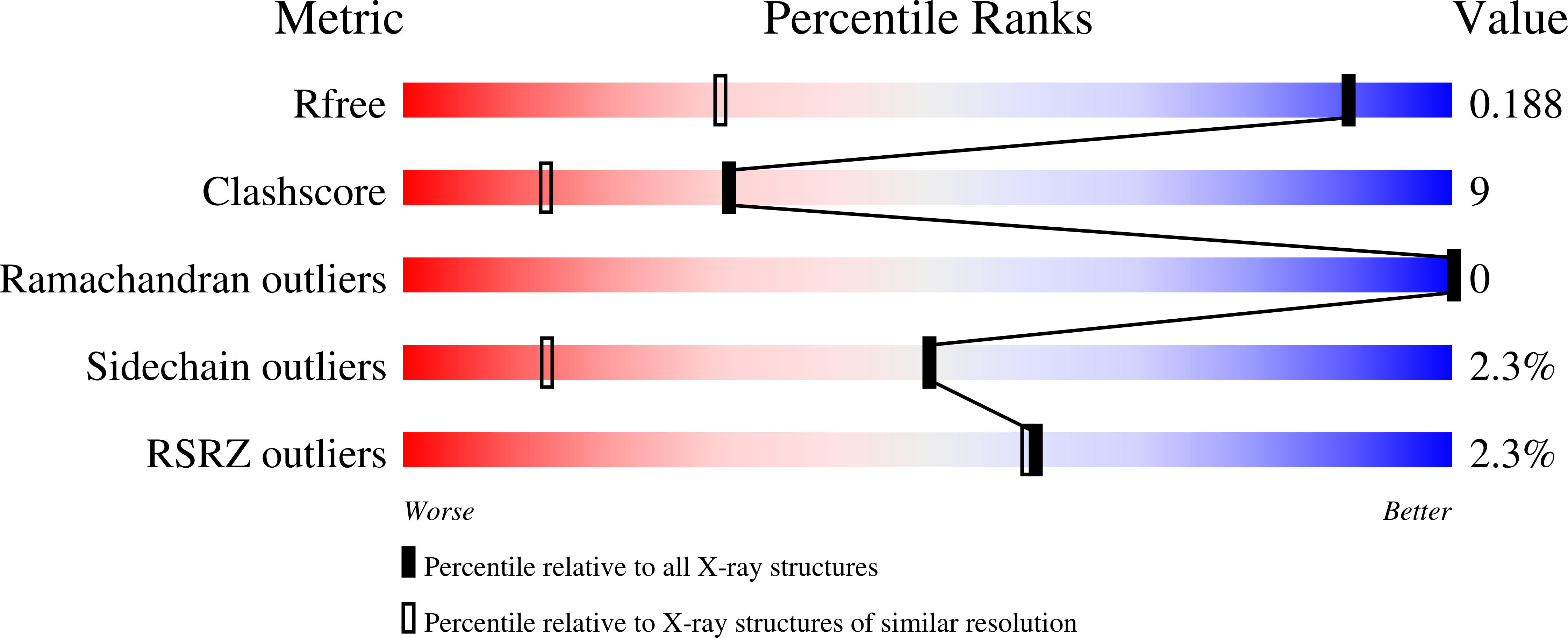
3RJ7 - PubMed Abstract:
Enhanced receptor selectivity: carbonic anhydrase inhibitors are relevant for both cancer diagnosis and therapy. Combining non-radioactive Re compounds with their radioactive (99m)Tc homologs enables the use of identical molecules for therapy and imaging (theragnostic). The syntheses and in vitro evaluation of [(Cp-R)M(CO)(3)] (Cp=cyclopentadienyl, M=Re, (99m)Tc) with R being a highly potent carbonic-anhydrase-targeting vector is reported.
Organizational Affiliation:
Institute of Inorganic Chemistry, University of Zurich, Switzerland.